Overview
Teaching: 40 min
Exercises: 30 minQuestions
How does CUDA programming with Numba work?
What CUDA features are available in Numba?
Objectives
Understand how to write CUDA programs using Numba.
Understand how Numba deals with CUDA threads.
Understand how Numba supports the CUDA memory models.
Numba supports CUDA GPU programming by directly compiling a restricted subset of Python code into CUDA kernels and device functions following the CUDA execution model.
One feature that significantly simplifies writing GPU kernels is that Numba makes it appear that the kernel has direct access to NumPy arrays. NumPy arrays that are supplied as arguments to the kernel are transferred between the CPU and the GPU automatically (although this can also be an issue).
Numba does not yet implement the full CUDA API, so some features are not available. However the features that are provided are enough to begin experimenting with writing GPU enable kernels. CUDA support in Numba is being actively developed, so eventually most of the features should be available.
Terminology
Several important terms in the topic of CUDA programming are listed here:
- host
- the CPU
- device
- the GPU
- host memory
- the system main memory
- device memory
- onboard memory on a GPU card
- kernel
- a GPU function launched by the host and executed on the device
- device function
- a GPU function executed on the device which can only be called from the device (i.e. from a kernel or another device function)
Device Management
It is possible to obtain a list of all the GPUs in the system using the following commands:
from numba import cuda
print(cuda.gpus)
If you do not have a CUDA-enabled GPU on your system, you will receive one of the following errors:
numba.cuda.cudadrv.error.CudaDriverError: CUDA initialized before forking
CudaSupportError: Error at driver init:
[3] Call to cuInit results in CUDA_ERROR_NOT_INITIALIZED:
numba.cuda.cudadrv.error.CudaDriverError: Error at driver init:
CUDA disabled by user:
If you do have a CUDA-enabled GPU on your system, you should see a message like:
<Managed Device 0>
If your machine has multiple GPUs, you might want to select which one to use. By default the CUDA driver selects the fastest GPU as the device 0, which is the default device used by Numba.
numba.cuda.select_device( device_id )
This creates a new CUDA context for the selected device_id. device_id should be the number of the device (starting from 0; the device order
is determined by the CUDA libraries). The context is associated with the current thread. Numba currently allows only one context per thread.
Using the CUDA simulator
If you don’t have a CUDA-enabled GPU (i.e. you received one of the error messages described previously), then you will need to use the CUDA simulator.
The simulator is enabled by setting the environment variable NUMBA_ENABLE_CUDASIM to 1.
Mac/Linux
Launch a terminal shell and type the commands:
export NUMBA_ENABLE_CUDASIM=1
Windows
Launch a CMD shell and type the commands:
SET NUMBA_ENABLE_CUDASIM=1
Now rerun the Device List command and check that you get the correct output.
Writing CUDA kernels
CUDA has an execution model unlike the traditional sequential model used for programming CPUs. In CUDA, the code you write will be executed by multiple threads at once (often hundreds or thousands). Your solution will be modeled by defining a thread hierarchy of grid, blocks, and threads.
Numba also exposes three kinds of GPU memory:
- global device memory
- shared memory
- local memory
For all but the simplest algorithms, it is important that you carefully consider how to use and access memory in order to minimize bandwidth requirements and contention.
NVIDIA recommends that programmers focus on following those recommendations to achieve the best performance:
- Find ways to parallelize sequential code
- Minimize data transfers between the host and the device
- Adjust kernel launch configuration to maximize device utilization
- Ensure global memory accesses are coalesced
- Minimize redundant accesses to global memory whenever possible
- Avoid different execution paths within the same warp
Kernel declaration
A kernel function is a GPU function that is meant to be called from CPU code. It has two fundamental characteristics:
- kernels cannot explicitly return a value; all result data must be written to an array passed to the function (if computing a scalar, you will probably pass a one-element array);
- kernels explicitly declare their thread hierarchy when called: i.e. the number of thread blocks and the number of threads per block (note that while a kernel is compiled once, it can be called multiple times with different block sizes or grid sizes).
from numba import cuda
@cuda.jit
def my_kernel(io_array):
"""
Code for kernel.
"""
# code here
Kernel invocation
A kernel is typically launched in the following way:
import numpy
# Create the data array - usually initialized some other way
data = numpy.ones(256)
# Set the number of threads in a block
threadsperblock = 32
# Calculate the number of thread blocks in the grid
blockspergrid = (data.size + (threadsperblock - 1)) // threadsperblock
# Now start the kernel
my_kernel[blockspergrid, threadsperblock](data)
# Print the result
print(data)
There are two main steps:
- Instantiate the kernel proper, by specifying a number of blocks per grid and a number of threads per block. The product of the two will give the total number of threads launched. Kernel instantiation is done by taking the compiled kernel function and indexing it with a tuple of integers.
- Running the kernel, by passing it the input array (and any separate output arrays if necessary). By default, running a kernel is synchronous: the function returns when the kernel has finished executing and the data is synchronized back.
Choosing the block size
The two-level thread hierarchy is important for the following reasons:
- On the software side, the block size determines how many threads share a given area of shared memory.
- On the hardware side, the block size must be large enough for full occupation of execution units; recommendations can be found in the CUDA C Programming Guide.
The block size you choose depends on a range of factors, including:
- The size of the data array
- The size of the shared mempory per block (e.g. 64KB)
- The maximum number of threads per block supported by the hardware (e.g. 512 or 1024)
- The maximum number of threads per multiprocessor (MP) (e.g. 2048)
- The maximum number of blocks per MP (e.g. 32)
- The number of threads that can be executed concurrently (a “warp” i.e. 32)
The execution of threads in a warp has a big effect on the computational throughput. If all threads in a warp are executing the same instruction then they can all be executed in parallel. But if one or more threads is executing a different instruction, the warp has to be split into groups of threads, and these groups execute serially.
Rules of thumb for threads per block:
- Should be a round multiple of the warp size (32)
- A good place to start is 128-512 but benchmarking is required to determine the optimal value.
Each streaming multiprocessor (SP) on the GPU must have enough active warps to achieve maximum throughput. In other words, the blocksize is usually selected to maximize the “occupancy”. See the CUDA Occupancy Calculator spreadsheet for more details.
Thread positioning
When running a kernel, the kernel function’s code is executed by every thread once. It therefore has to know which thread it is in, in order to know which array element(s) it is responsible for. More complex algorithms may define more complex responsibilities, but the underlying principle is the same.
To help deal with multi-dimensional arrays, CUDA allows you to specify multi-dimensional blocks and grids. In the example above, you could
make blockspergrid and threadsperblock tuples of one, two or three integers. Compared to 1-dimensional declarations of equivalent sizes,
this doesn’t change anything to the efficiency or behaviour of generated code, but can help you write your algorithms in a more natural way.
One way is for the thread to determines its position in the grid and block and manually compute the corresponding array position:
@cuda.jit
def my_kernel(io_array):
# Thread id in a 1D block
tx = cuda.threadIdx.x
# Block id in a 1D grid
ty = cuda.blockIdx.x
# Block width, i.e. number of threads per block
bw = cuda.blockDim.x
# Compute flattened index inside the array
pos = tx + ty * bw
if pos < io_array.size: # Check array boundaries
io_array[pos] *= 2 # do the computation
Note: Unless you are sure the block size and grid size is a divisor of your array size, you must check boundaries as shown above.
The following special objects are provided by the CUDA backend for the sole purpose of knowing the geometry of the thread hierarchy and the position of the current thread within that geometry:
numba.cuda.threadIdx- The thread indices in the current thread block. For 1-dimensional blocks, the index (given by the x attribute) is an integer spanning the range from 0 tonumba.cuda.blockDim- 1. A similar rule exists for each dimension when more than one dimension is used.numba.cuda.blockDim- The shape of the block of threads, as declared when instantiating the kernel. This value is the same for all threads in a given kernel, even if they belong to different blocks (i.e. each block is “full”).numba.cuda.blockIdx- The block indices in the grid of threads launched a kernel. For a 1-dimensional grid, the index (given by thexattribute) is an integer spanning the range from 0 tonumba.cuda.gridDim- 1. A similar rule exists for each dimension when more than one dimension is used.numba.cuda.gridDim- The shape of the grid of blocks, i.e. the total number of blocks launched by this kernel invocation, as declared when instantiating the kernel.
These objects can be 1-, 2- or 3-dimensional, depending on how the kernel was invoked. To access the value at each dimension, use the x, y and z
attributes of these objects, respectively.
Absolute positions
Simple algorithms will tend to always use thread indices in the same way as shown in the example above. Numba provides additional facilities to automate such calculations:
numba.cuda.grid(ndim)- Return the absolute position of the current thread in the entire grid of blocks.ndimshould correspond to the number of dimensions declared when instantiating the kernel. Ifndimis 1, a single integer is returned. Ifndimis 2 or 3, a tuple of the given number of integers is returned.numba.cuda.gridsize(ndim)- Return the absolute size (or shape) in threads of the entire grid of blocks.ndimhas the same meaning as ingrid()above.
Using these functions, the our example can become:
@cuda.jit
def my_kernel2(io_array):
pos = cuda.grid(1)
if pos < io_array.size:
io_array[pos] *= 2 # do the computation
Now we need to add the host code that calls the kernel. Notice that the grid computation when instantiating the kernel must still be done manually.
The complete program should be called cuda1.py and is shown below.
from __future__ import division
from numba import cuda
import numpy
import math
# CUDA kernel
@cuda.jit
def my_kernel(io_array):
pos = cuda.grid(1)
if pos < io_array.size:
io_array[pos] *= 2 # do the computation
# Host code
data = numpy.ones(256)
threadsperblock = 256
blockspergrid = math.ceil(data.shape[0] / threadsperblock)
my_kernel[blockspergrid, threadsperblock](data)
print(data)
Now run the program with the following command (remember to set NUMBA_ENABLE_CUDASIM=1 if you don’t have an NVIDIA GPU):
python cuda1.py
You should see the following output:
[ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.
2. 2. 2. 2.]
Challenge
Assuming that
x, y = cuda.grid(2)returns the thread indices, rewrite the kernel to work ifio_arrayis a 2-dimensional array. Make sure that you verify thatxandyare within the bounds of the array (useio_array.shapeinstead ofio_array.size). Run the kernel using the following host program.from __future__ import division from numba import cuda import numpy import math @cuda.jit def my_kernel_2D(io_array): x, y = cuda.grid(2) ### YOUR SOLUTION HERE data = numpy.ones((16, 16)) threadsperblock = (16, 16) blockspergrid_x = math.ceil(data.shape[0] / threadsperblock[0]) blockspergrid_y = math.ceil(data.shape[1] / threadsperblock[1]) blockspergrid = (blockspergrid_x, blockspergrid_y) my_kernel_2D[blockspergrid, threadsperblock](data) print(data)Here is the output you should see if the kernel is correct:
[[ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.] [ 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2. 2.]]
Memory management
Numba has been automatically transferring the NumPy arrays to the device when you invoke the kernel. However, it can only do so conservatively by always transferring the device memory back to the host when a kernel finishes. To avoid the unnecessary transfer for read-only arrays, it is possible to manually control the transfer.
device_array = cuda.device_array( shape )
Allocate an empty device ndarray. Similar to numpy.empty().
device_array = cuda.to_device( array )
Allocate and transfer a NumPy ndarray to the device.
A more complex example: matrix multiplication
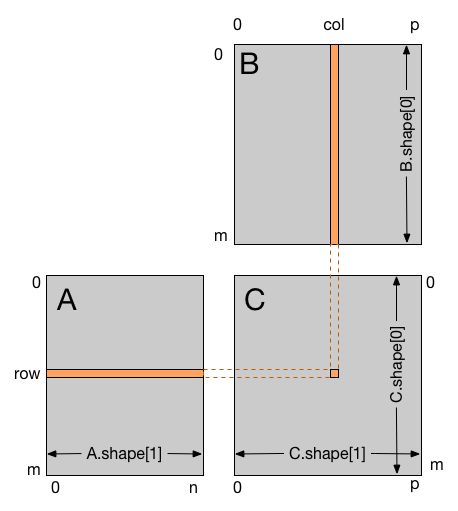
The following code sample is a straightforward implementation of matrix multiplication for matrices where each thread reads one row of A and
one column of B and computes the corresponding element of C. For input arrays where A.shape == (m, n) and B.shape == (n, p) then
the result shape will be C.shape = (m, p).
@cuda.jit
def matmul(A, B, C):
"""Perform matrix multiplication of C = A * B
"""
row, col = cuda.grid(2)
if row < C.shape[0] and col < C.shape[1]:
tmp = 0.
for k in range(A.shape[1]):
tmp += A[row, k] * B[k, col]
C[row, col] = tmp
The host code must create and initiliaze the A and B arrays, then move them to the device. Next, it must allocate space on the device for the result
array. Once the kernel has completed, the result array must be copied back to the host so that it can be displayed.
Create a program called cuda2.py using the code below to see how the kernel works for the input arrays. Notice that the number of threads per block
and blocks per grid is not really important, other than to ensure that there are enough threads to complete the calculation.
from __future__ import division
from numba import cuda
import numpy
import math
# CUDA kernel
@cuda.jit
def matmul(A, B, C):
"""Perform matrix multiplication of C = A * B
"""
row, col = cuda.grid(2)
if row < C.shape[0] and col < C.shape[1]:
tmp = 0.
for k in range(A.shape[1]):
tmp += A[row, k] * B[k, col]
C[row, col] = tmp
# Host code
# Initialize the data arrays
A = numpy.full((24, 12), 3, numpy.float) # matrix containing all 3's
B = numpy.full((12, 22), 4, numpy.float) # matrix containing all 4's
# Copy the arrays to the device
A_global_mem = cuda.to_device(A)
B_global_mem = cuda.to_device(B)
# Allocate memory on the device for the result
C_global_mem = cuda.device_array((24, 22))
# Configure the blocks
threadsperblock = (16, 16)
blockspergrid_x = int(math.ceil(A.shape[0] / threadsperblock[0]))
blockspergrid_y = int(math.ceil(B.shape[1] / threadsperblock[1]))
blockspergrid = (blockspergrid_x, blockspergrid_y)
# Start the kernel
matmul[blockspergrid, threadsperblock](A_global_mem, B_global_mem, C_global_mem)
# Copy the result back to the host
C = C_global_mem.copy_to_host()
print(C)
A problem with this code is that each thread is reading from the global memory containing the copies of A and B. In fact, the A global memory
is read B.shape[1] times and the B global memory is read A.shape[0] times. Since global memory is fairly slow, this results in an inefficient
use of the GPU.
Shared memory and thread synchronization
A limited amount of shared memory can be allocated on the device to speed up access to data, when necessary. That memory will be shared (i.e. both readable and writable) amongst all threads belonging to a given block and has faster access times than regular device memory. It also allows threads to cooperate on a given solution. You can think of it as a manually-managed data cache.
The function to create a shared memory array is:
shared_array = cuda.shared.array(shape,type)
shape- is either an integer or a tuple of integers representing the array’s dimensions.
type- is a Numba type of the elements needing to be stored in the array
The memory is allocated once for the duration of the kernel, unlike traditional dynamic memory management.
Because the shared memory is a limited resource, it is often necessary to preload a small block at a time from the input arrays. All the threads then need to wait until everyone has finished preloading before doing the computation on the shared memory.
Synchronization is then required again after the computation to ensure all threads have finished with the data in shared memory before overwriting it in the next loop iteration.
The function to synchronized threads is:
cuda.syncthreads()
This function will synchronize all threads in the same thread block. This function implements the same pattern as barriers in traditional
multi-threaded programming and the MPI.Barrier() function. The program will wait until all threads in the block call the function, at
which point it returns control to all its callers.
Improved matrix multiplication
The following example shows how shared memory can be used when performing matrix multiplication.
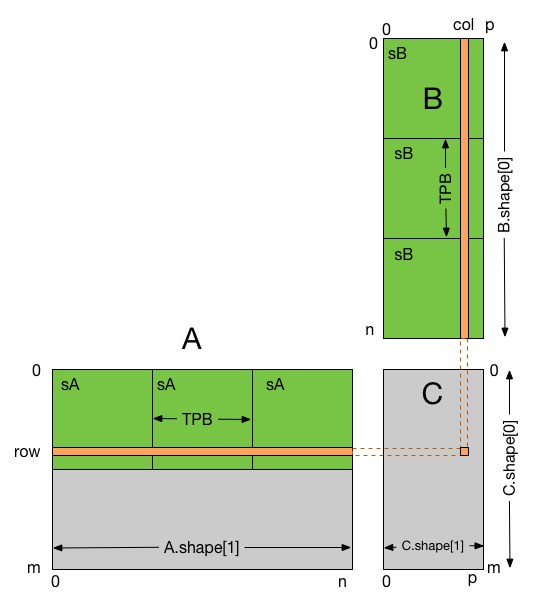
In this example, each thread block is responsible for computing a square sub-matrix of C and each thread for computing an element of the sub-matrix.
The sub-matrix is equal to the product of a square sub-matrix of A (sA) and a square sub-matrix of B (sB). In order to fit into the device
resources, the two input matrices are divided into as many square sub-matrices of dimension TPB as necessary, and the result computed as the
sum of the products of these square sub-matrices.
Each product is performed by first loading sA and sB from global memory to shared memory, with one thread loading each element of each sub-matrix.
Once sA and sB have been loaded, each thread accumulates the result into a register (tmp). Once all the products have been calculated, the
results are written to the matrix C in global memory.
By blocking the computation this way, we can reduce the number of global memory accesses since A is now only read B.shape[1] / TPB times and B
is read A.shape[0] / TPB times.
from __future__ import division
from numba import cuda, float32
import numpy
import math
# Controls threads per block and shared memory usage.
# The computation will be done on blocks of TPBxTPB elements.
TPB = 16
@cuda.jit
def fast_matmul(A, B, C):
"""
Perform matrix multiplication of C = A * B
Each thread computes one element of the result matrix C
"""
# Define an array in the shared memory
# The size and type of the arrays must be known at compile time
sA = cuda.shared.array(shape=(TPB, TPB), dtype=float32)
sB = cuda.shared.array(shape=(TPB, TPB), dtype=float32)
x, y = cuda.grid(2)
tx = cuda.threadIdx.x
ty = cuda.threadIdx.y
if x >= C.shape[0] and y >= C.shape[1]:
# Quit if (x, y) is outside of valid C boundary
return
# Each thread computes one element in the result matrix.
# The dot product is chunked into dot products of TPB-long vectors.
tmp = 0.
for i in range(int(A.shape[1] / TPB)):
# Preload data into shared memory
sA[tx, ty] = A[x, ty + i * TPB]
sB[tx, ty] = B[tx + i * TPB, y]
# Wait until all threads finish preloading
cuda.syncthreads()
# Computes partial product on the shared memory
for j in range(TPB):
tmp += sA[tx, j] * sB[j, ty]
# Wait until all threads finish computing
cuda.syncthreads()
C[x, y] = tmp
# The data array
A = numpy.full((TPB*2, TPB*3), 3, numpy.float) # [32 x 48] matrix containing all 3's
B = numpy.full((TPB*3, TPB*1), 4, numpy.float) # [48 x 16] matrix containing all 4's
A_global_mem = cuda.to_device(A)
B_global_mem = cuda.to_device(B)
C_global_mem = cuda.device_array((TPB*2, TPB*1)) # [32 x 16] matrix result
# Configure the blocks
threadsperblock = (TPB, TPB)
blockspergrid_x = int(math.ceil(A.shape[0] / threadsperblock[1]))
blockspergrid_y = int(math.ceil(B.shape[1] / threadsperblock[0]))
blockspergrid = (blockspergrid_x, blockspergrid_y)
# Start the kernel
fast_matmul[blockspergrid, threadsperblock](A_global_mem, B_global_mem, C_global_mem)
res = C_global_mem.copy_to_host()
print(res)
Create a new program called cuda3.py using your new kernel and the host program to verify that it works correctly.
Key Points
Numba provides an easy way to write CUDA programs.
Many CUDA features are provided by Numba.
Some of the low level CUDA API features are not supported by Numba.